readme.md 18 KB
Image Features Extraction Package
This package allows the fast extraction and classification of features from a set of images.
Package documentation
Tutorial
This Python package allows the fast extraction and classification of features from a set of images. The resulting data frame can be used as training and testing set for machine learning classifier.
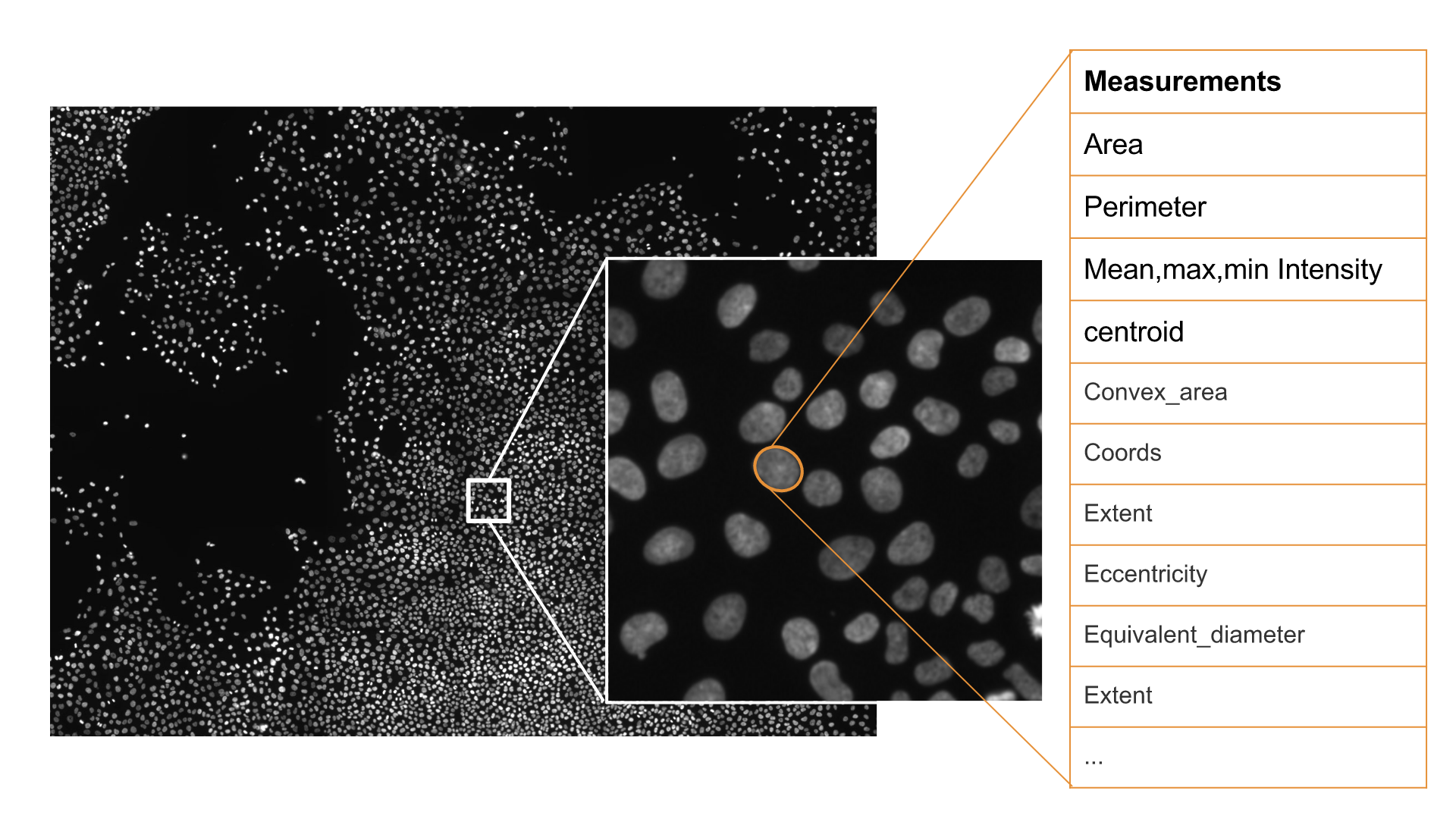
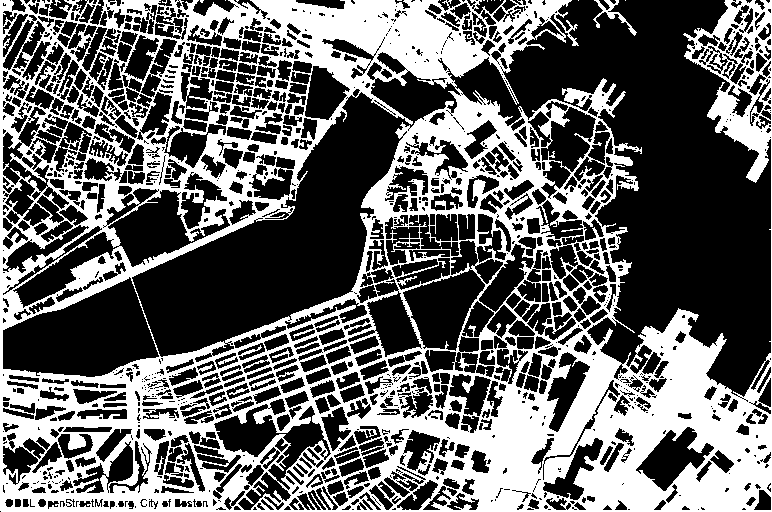
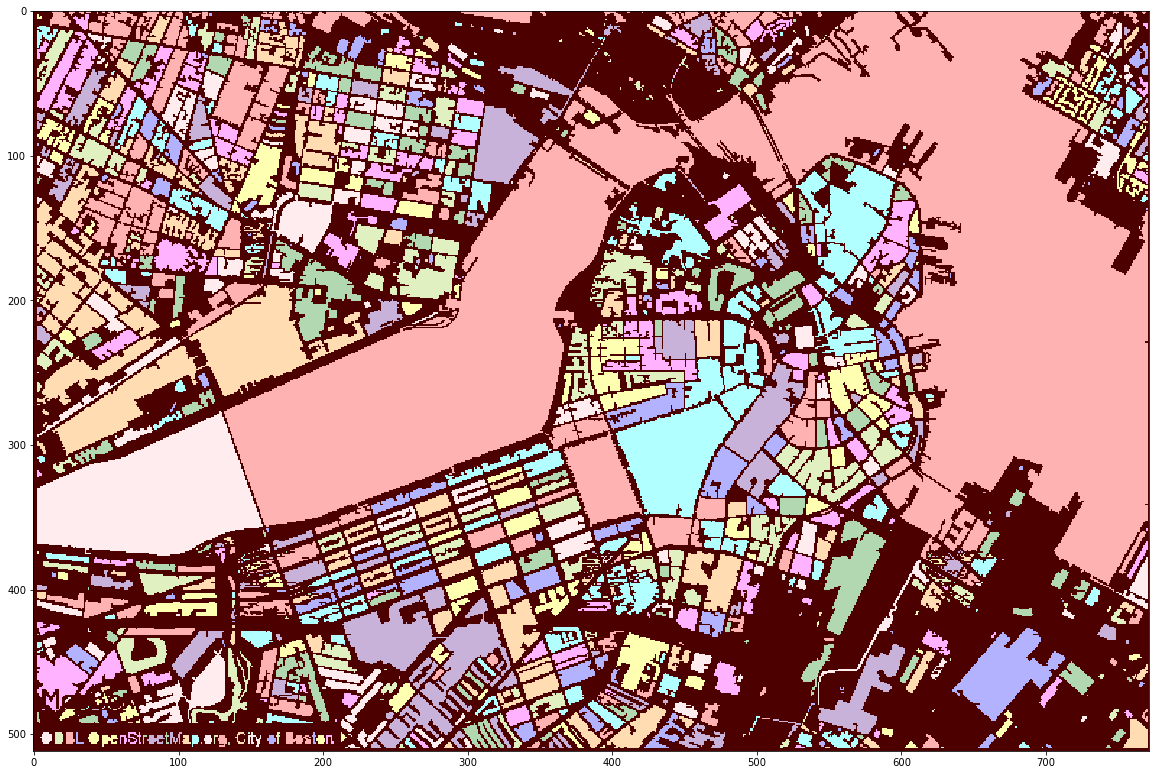
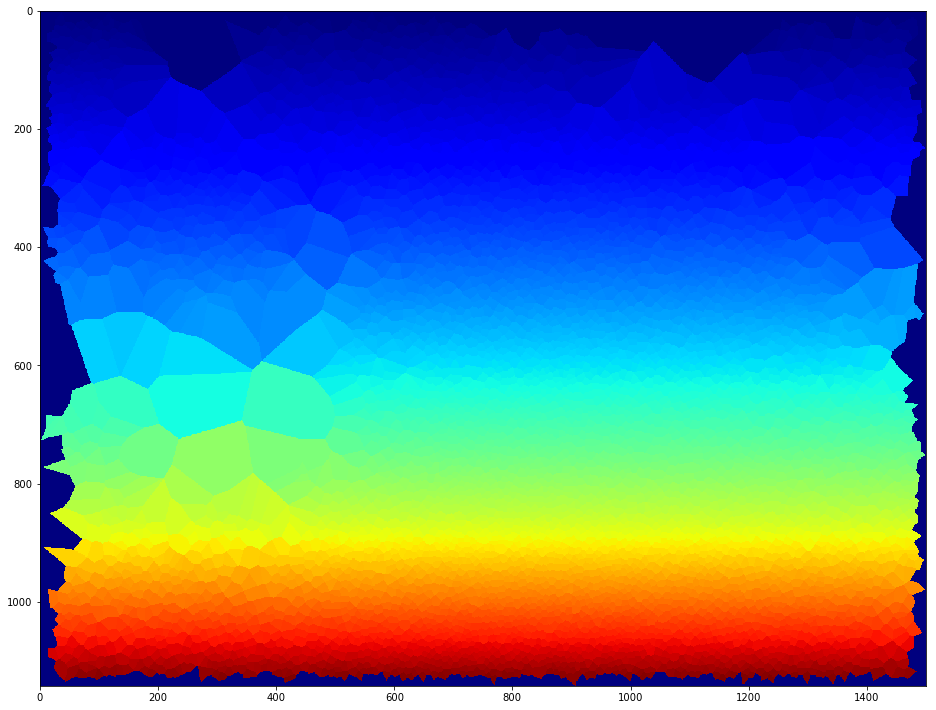
This package was originally developed to extract measurements of single cell nuclei from microscopy images (see figure above). The package can be used to extract features from any set of images for a variety of applications. Below it is shown a map of Boston used for city density and demographic models.
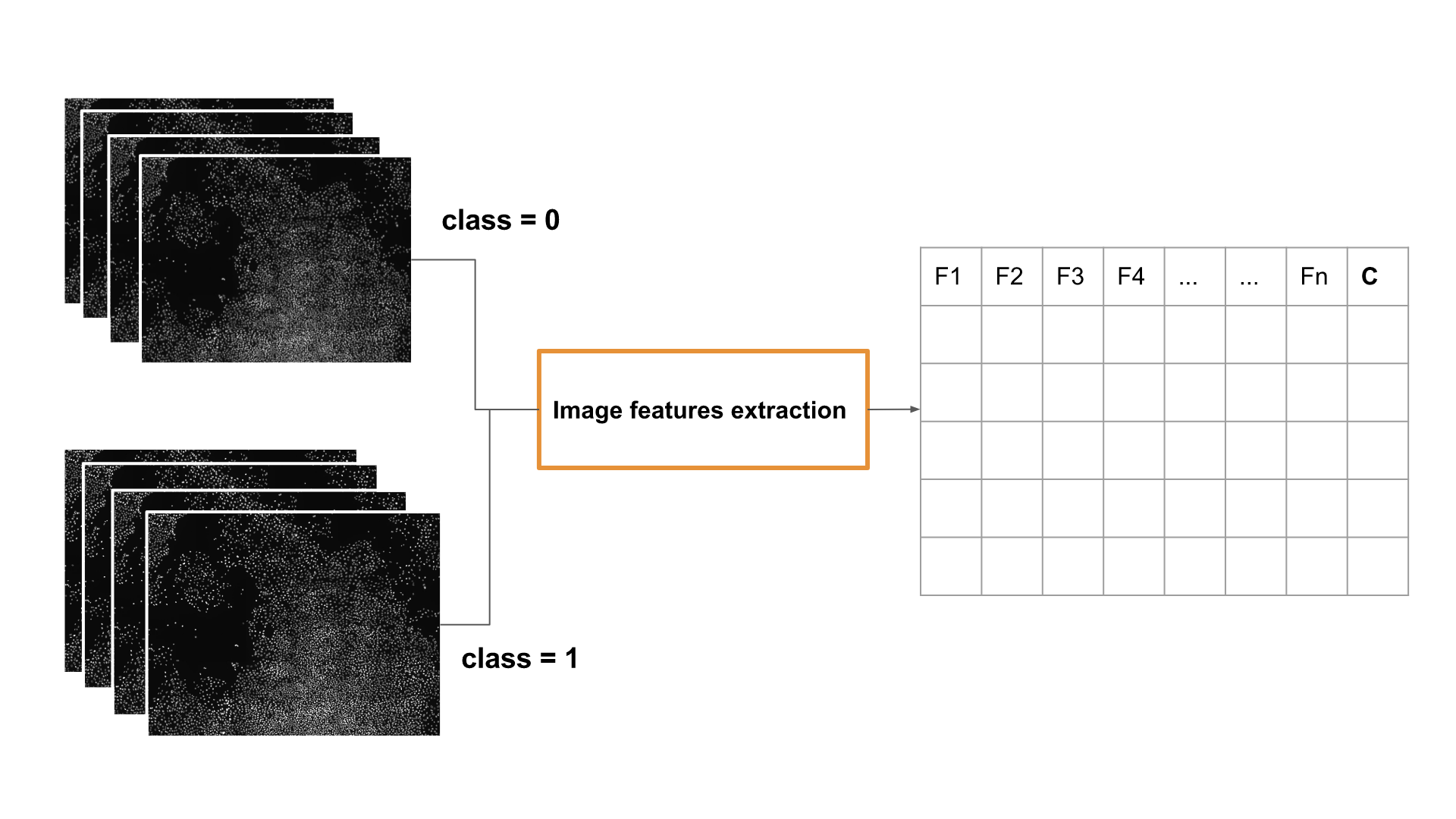
Features extraction for spatial classification of images
The image below shows a possible workflow for image feature extraction: two sets of images with different classification labels are used to produce two data sets for training and testing a classifier
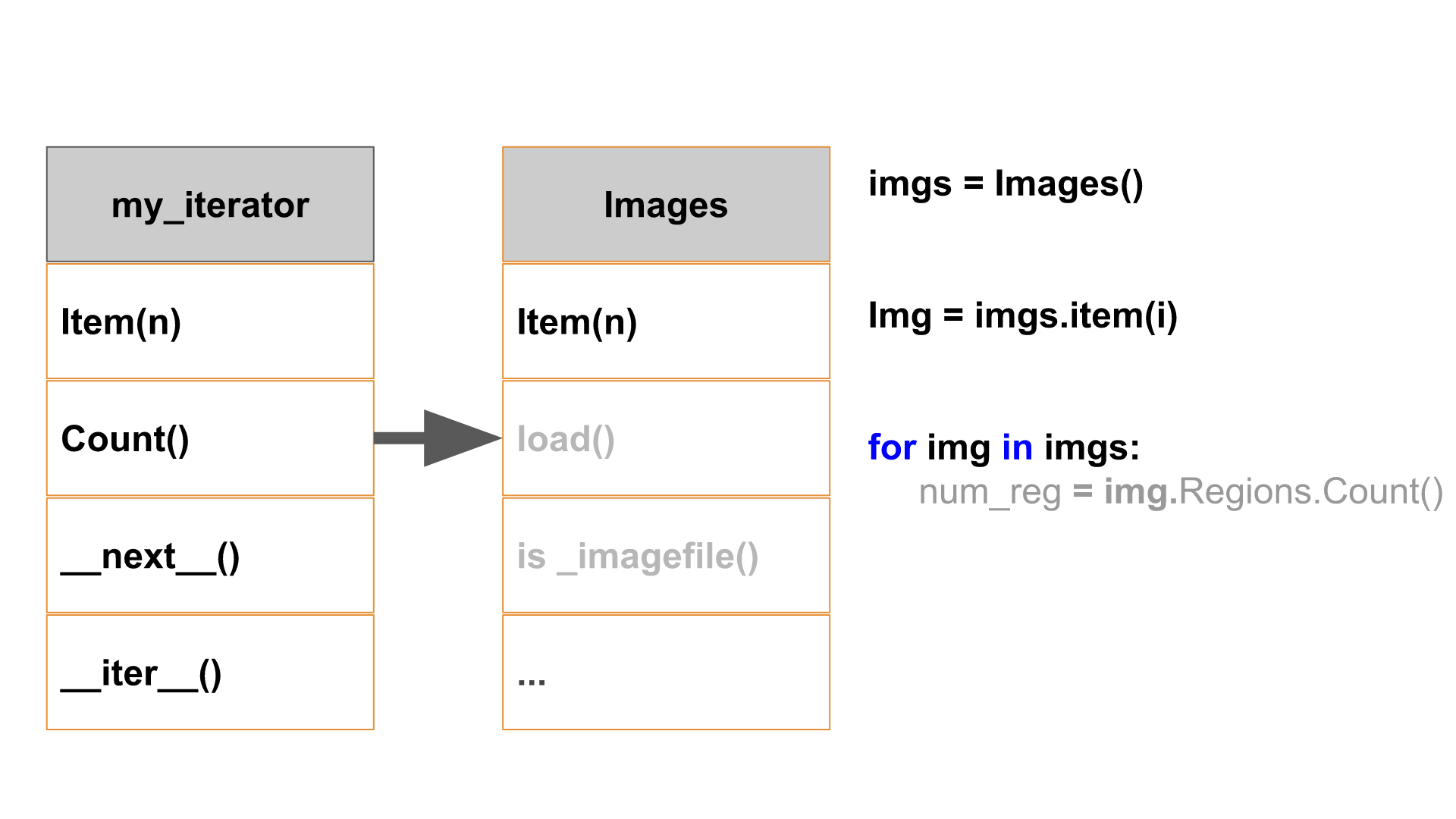
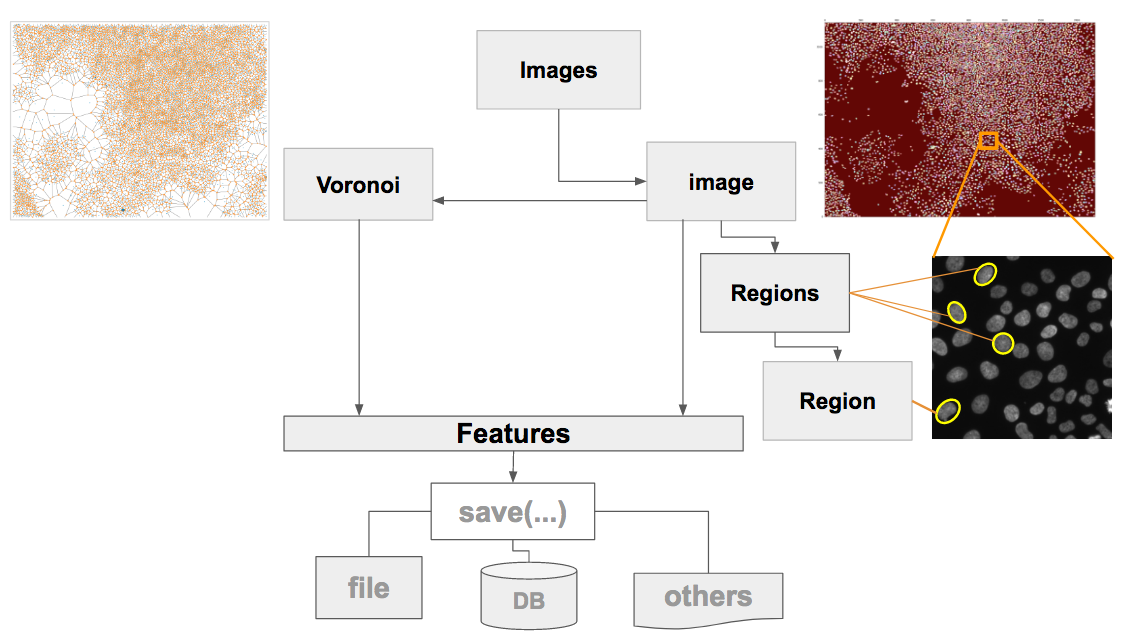
An example of Collection-object and Iterator implementation
The object 'Image' includes the function Voronoi(), which returns the object Voronoi of my package Voronoi_Features. The Voronoi object can be used to measure the voronoi tassels of each image regions. It includes >30 measurements. Below an example of voronoi diagrams from the image shown above
Image features extraction for city density and demographic analysis modelling
Create the Images root object and laod the images contained in the folder
% matplotlib inline
import matplotlib.pyplot as plt
import image_features_extraction.Images as fe
IMGS = fe.Images('../images/CITY')
IMG = IMGS.item(0)
print(IMG.file_name())
fig, ax = plt.subplots(figsize=(20, 20))
ax.imshow(IMGS.item(0).get_image_segmentation())
../images/CITY/Boston_Center.tif
<matplotlib.image.AxesImage at 0x11f3e2400>
features = IMG.features(['label', 'area','perimeter', 'centroid', 'moments'])
df2 = features.get_dataframe()
df2.head()
| id | label | area | perimeter | centroid_x | centroid_y | moments | |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 44 | 4 | 4.000000 | 2.500000 | 122.500000 | [[4.0, 2.0, 2.0, 2.0], [2.0, 1.0, 1.0, 1.0], [... |
| 1 | 1 | 45 | 6 | 5.207107 | 4.333333 | 3.833333 | [[6.0, 8.0, 14.0, 26.0], [5.0, 8.0, 14.0, 26.0... |
| 2 | 2 | 46 | 64 | 36.556349 | 7.718750 | 34.015625 | [[64.0, 302.0, 1862.0, 13058.0], [385.0, 1857.... |
| 3 | 3 | 47 | 29 | 23.520815 | 6.517241 | 146.689655 | [[29.0, 102.0, 476.0, 2580.0], [78.0, 305.0, 1... |
| 4 | 4 | 48 | 165 | 62.355339 | 10.121212 | 460.951515 | [[165.0, 1175.0, 10225.0, 99551.0], [1807.0, 1... |
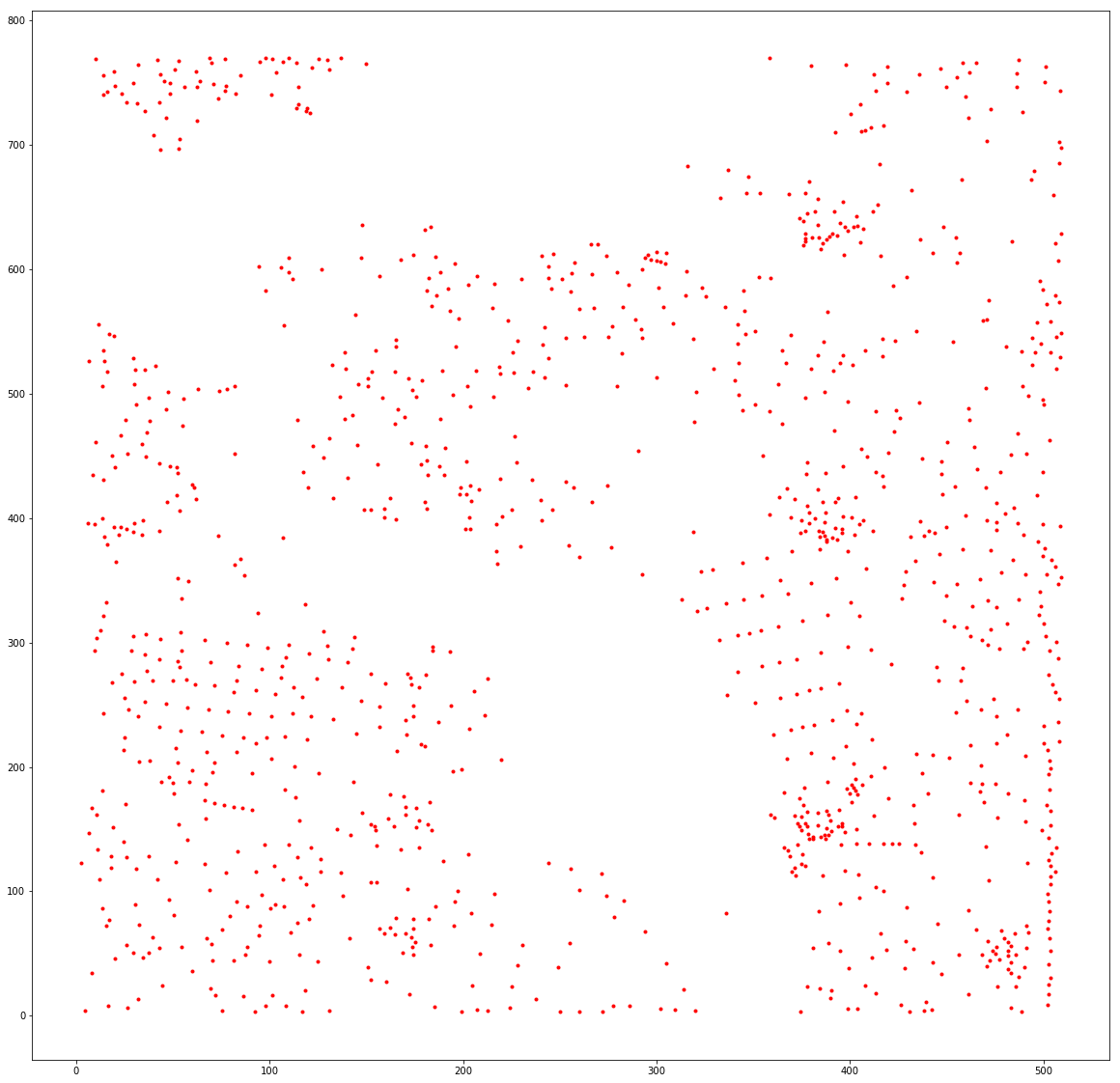
# SHOW THE FOUND CENTROIDS
fig, ax = plt.subplots(figsize=(20, 20))
plt.plot(df2.centroid_x,df2.centroid_y,'.r' )
[<matplotlib.lines.Line2D at 0x119b1ea58>]
h = plt.hist(df2.area,100)
Image features extraction for cellular spatial analysis
Images show cell nuclei
% matplotlib inline
import matplotlib.pyplot as plt
import image_features_extraction.Images as fe
IMGS = fe.Images('../images/CA/1')
# the iterator at work ...
for IMG in IMGS:
print(IMG.file_name())
../images/CA/1/ORG_8bit.tif
../images/CA/1/ORG_bin.tif
fig, ax = plt.subplots(figsize=(20, 20))
ax.imshow(IMGS.item(1).get_image_segmentation())
<matplotlib.image.AxesImage at 0x11ab282b0>
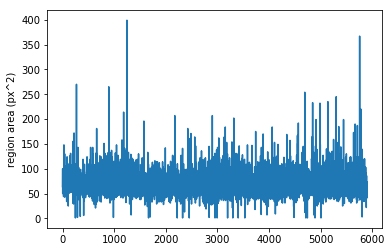
An example of measurement and visualization of a property, e.g., area
IMG = IMGS.item(1)
REGS = IMG.regions()
areas = REGS.prop_values('area')
plt.plot(areas)
plt.ylabel('region area (px^2)')
<matplotlib.text.Text at 0x11f38b048>
h = plt.hist(df2.area,100)
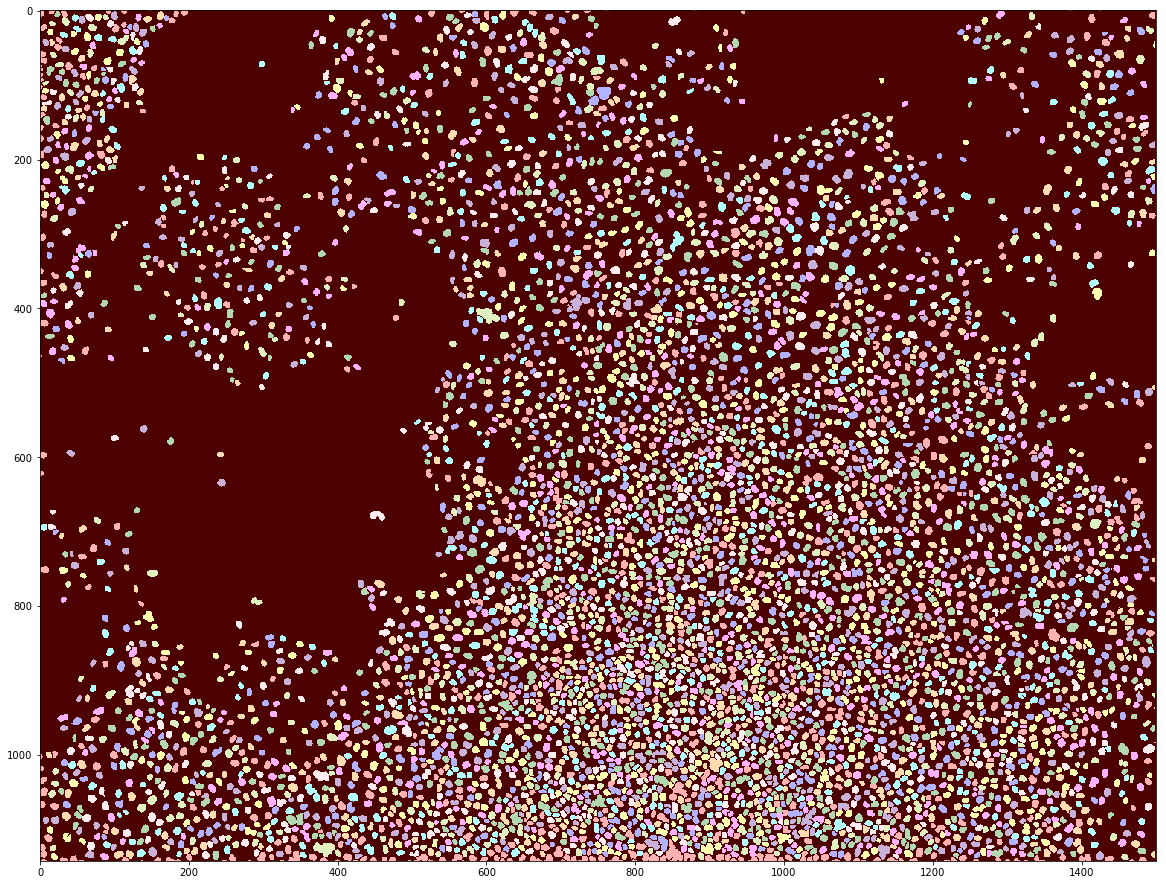
VORONOI FEATURES
vor = IMG.Voronoi()
vor = IMG.Voronoi()
IMG_VOR = vor.get_voronoi_map()
fig = plt.figure(figsize=(20,20))
plt.imshow(IMG_VOR, cmap=plt.get_cmap('jet'))
<matplotlib.image.AxesImage at 0x11d228e48>
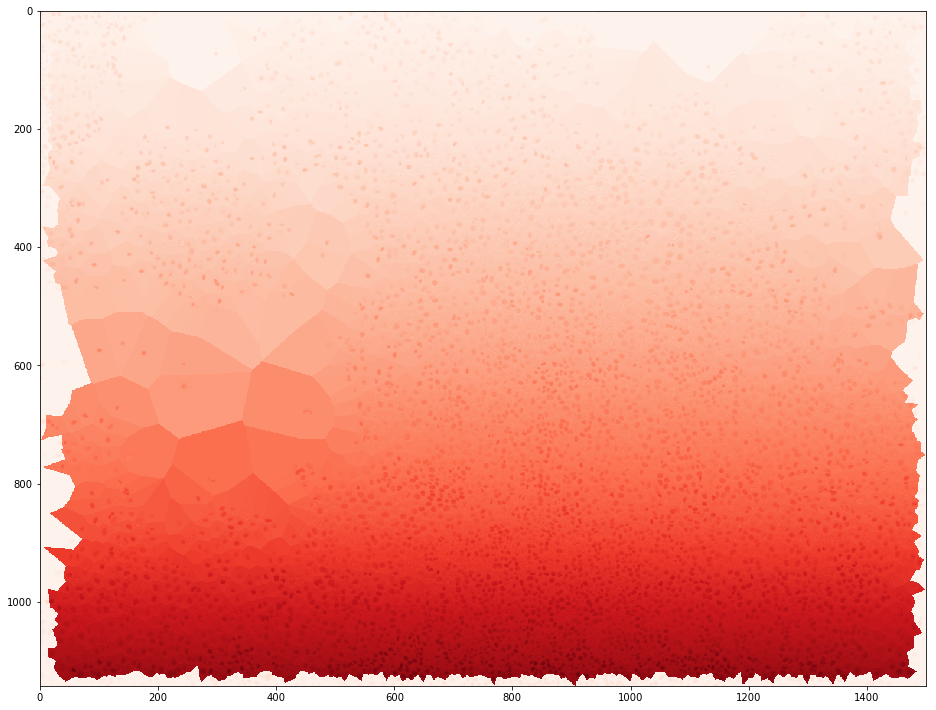
i1 = IMGS.item(0).get_image_segmentation()
i2 = vor.get_voronoi_map()
i3 = i1[:,:,0] + i2/1000
fig = plt.figure(figsize=(yinch,xinch))
plt.imshow(i3, cmap=plt.get_cmap('Reds'))
<matplotlib.image.AxesImage at 0x11ebbd6d8>
Feature from the image only
features1 = IMG.features(['area','perimeter','centroid','bbox', 'eccentricity'])
features1.get_dataframe().head()
| id | area | perimeter | centroid_x | centroid_y | bbox | eccentricity | |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 4 | 4.000000 | 2.500000 | 122.500000 | (2, 122, 4, 124) | 0.000000 |
| 1 | 1 | 6 | 5.207107 | 4.333333 | 3.833333 | (3, 3, 6, 6) | 0.738294 |
| 2 | 2 | 64 | 36.556349 | 7.718750 | 34.015625 | (3, 28, 14, 39) | 0.410105 |
| 3 | 3 | 29 | 23.520815 | 6.517241 | 146.689655 | (3, 144, 11, 151) | 0.736301 |
| 4 | 4 | 165 | 62.355339 | 10.121212 | 460.951515 | (3, 450, 19, 471) | 0.718935 |
Features from the voronoi diagram only
features2 = vor.features(['area','perimeter','centroid','bbox', 'eccentricity'])
features2.get_dataframe().head()
| id | voro_area | voro_perimeter | voro_centroid | voro_bbox | voro_eccentricity | |
|---|---|---|---|---|---|---|
| 0 | 24 | 314 | 71.112698 | (13.9203821656, 407.257961783) | (2, 395, 25, 416) | 0.502220 |
| 1 | 33 | 365 | 78.526912 | (18.2, 481.273972603) | (2, 473, 32, 491) | 0.861947 |
| 2 | 71 | 343 | 94.911688 | (17.8717201166, 723.320699708) | (3, 706, 30, 740) | 0.955651 |
| 3 | 32 | 161 | 50.662951 | (15.7701863354, 450.565217391) | (5, 445, 24, 460) | 0.738073 |
| 4 | 46 | 160 | 50.591883 | (15.8625, 516.75) | (5, 511, 24, 524) | 0.782348 |
Merge features from the image + the voronoi diagram
features3 = features1.merge(features2, how_in='inner')
features3.get_dataframe().head()
| id | area | perimeter | centroid_x | centroid_y | bbox | eccentricity | voro_area | voro_perimeter | voro_centroid | voro_bbox | voro_eccentricity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 8 | 147 | 95.041631 | 18.843537 | 151.149660 | (5, 146, 34, 157) | 0.967212 | 257 | 67.355339 | (22.2762645914, 152.482490272) | (12, 143, 36, 162) | 0.799861 |
| 1 | 15 | 485 | 279.260931 | 25.649485 | 170.092784 | (8, 155, 40, 188) | 0.618654 | 447 | 80.325902 | (29.0604026846, 169.451901566) | (17, 157, 42, 185) | 0.558628 |
| 2 | 17 | 114 | 69.562446 | 20.061404 | 747.701754 | (8, 739, 33, 753) | 0.960308 | 73 | 31.798990 | (20.1369863014, 748.931506849) | (14, 744, 26, 754) | 0.530465 |
| 3 | 18 | 106 | 48.556349 | 17.990566 | 119.075472 | (9, 114, 28, 125) | 0.810733 | 151 | 48.763456 | (18.2185430464, 117.688741722) | (10, 109, 25, 124) | 0.756768 |
| 4 | 21 | 2 | 0.000000 | 9.500000 | 395.000000 | (9, 395, 11, 396) | 1.000000 | 63 | 33.349242 | (10.0158730159, 392.698412698) | (6, 387, 15, 400) | 0.742086 |
Add class name and value
features3.set_class_name('class')
features3.set_class_value('test_class_val')
features3.get_dataframe(include_class=True).head()
| id | area | perimeter | centroid_x | centroid_y | bbox | eccentricity | voro_area | voro_perimeter | voro_centroid | voro_bbox | voro_eccentricity | class | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 8 | 147 | 95.041631 | 18.843537 | 151.149660 | (5, 146, 34, 157) | 0.967212 | 257 | 67.355339 | (22.2762645914, 152.482490272) | (12, 143, 36, 162) | 0.799861 | test_class_val |
| 1 | 15 | 485 | 279.260931 | 25.649485 | 170.092784 | (8, 155, 40, 188) | 0.618654 | 447 | 80.325902 | (29.0604026846, 169.451901566) | (17, 157, 42, 185) | 0.558628 | test_class_val |
| 2 | 17 | 114 | 69.562446 | 20.061404 | 747.701754 | (8, 739, 33, 753) | 0.960308 | 73 | 31.798990 | (20.1369863014, 748.931506849) | (14, 744, 26, 754) | 0.530465 | test_class_val |
| 3 | 18 | 106 | 48.556349 | 17.990566 | 119.075472 | (9, 114, 28, 125) | 0.810733 | 151 | 48.763456 | (18.2185430464, 117.688741722) | (10, 109, 25, 124) | 0.756768 | test_class_val |
| 4 | 21 | 2 | 0.000000 | 9.500000 | 395.000000 | (9, 395, 11, 396) | 1.000000 | 63 | 33.349242 | (10.0158730159, 392.698412698) | (6, 387, 15, 400) | 0.742086 | test_class_val |
To measure intensity from image regions
The example below shows how to associate a grayscale image to a binary one for intensity measurement. The package uses intenally a very simple segmentation algorithm based on an Otsu Thresholding method for segmentation of binary images. The goal of the package is not to segment images but to measure their segmented features. The correct way to use this package is by using as input pre-segmented binary images and if intensity measurement are needed to associate the original grayscale image.
IMG = IMGS.item(1)
IMG.set_image_intensity(IMGS.item(0))
features = IMG.features(['label', 'area','perimeter', 'centroid', 'moments','mean_intensity'])
df = features.get_dataframe()
df.head()
| id | label | area | perimeter | centroid_x | centroid_y | moments | mean_intensity | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 22 | 64 | 28.278175 | 5.468750 | 584.375000 | [[64.0, 286.0, 1630.0, 10366.0], [280.0, 1223.... | 170.078125 |
| 1 | 1 | 23 | 86 | 33.556349 | 6.418605 | 621.546512 | [[86.0, 466.0, 3268.0, 25726.0], [391.0, 2067.... | 139.127907 |
| 2 | 2 | 24 | 100 | 35.556349 | 5.720000 | 1290.330000 | [[100.0, 472.0, 2988.0, 21442.0], [533.0, 2238... | 99.360000 |
| 3 | 3 | 25 | 50 | 24.142136 | 5.600000 | 23.040000 | [[50.0, 180.0, 846.0, 4458.0], [202.0, 699.0, ... | 181.940000 |
| 4 | 4 | 26 | 80 | 31.556349 | 7.325000 | 99.462500 | [[80.0, 426.0, 2894.0, 21846.0], [357.0, 1969.... | 157.675000 |
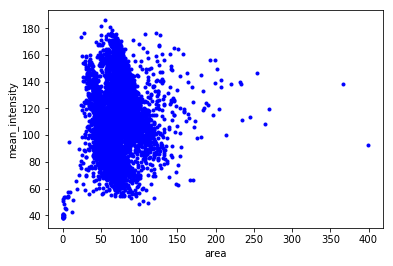
Plot area vs perimeter and area histogram
plt.plot(df.area, df.mean_intensity, '.b')
plt.xlabel('area')
plt.ylabel('mean_intensity')
<matplotlib.text.Text at 0x114a69908>
An example of how save measured features
This package includes the class Features for data managment layer, which is used to separate the business from the data layer and allow easy scalability of the data layer.
import image_features_extraction.Images as fe
IMGS = fe.Images('../images/EDGE')
storage_name = '../images/DB1.csv'
class_value = 1
for IMG in IMGS:
print(IMG.file_name())
REGS = IMG.regions()
FEATURES = REGS.features(['area','perimeter', 'extent', 'equivalent_diameter', 'eccentricity'], class_value=class_value)
FEATURES.save(storage_name, type_storage='file', do_append=True)
../images/EDGE/ca_1.tif
../images/EDGE/ca_2.tif
../images/EDGE/ca_3.tif
Pytest: Units test
!py.test
[1m============================= test session starts ==============================[0m
platform darwin -- Python 3.5.3, pytest-3.1.3, py-1.4.34, pluggy-0.4.0
rootdir: /Users/remi/Google Drive/INSIGHT PRJ/PRJ/Image-Features-Extraction, inifile:
collected 0 items [0m[1m
[0m
[1m[33m========================= no tests ran in 0.01 seconds =========================[0m